HYGENE: A Diffusion-based Hypergraph Generation Method
This blog post will summarize our paper, HYGENE: A Diffusion-based Hypergraph Generation Method [1], accepted at AAAI-25. To the best of our knowledge, this work introduces the first deep-learning based method for hypergraph generation. Code is available here.
TL;DR: We introduce a hypergraph generation method based on an “unzoom-zoom” principle. First, we compute increasingly coarse versions of a hypergraph (“unzoom”). Then, we train a model to reconstruct the original details from these simplified versions (“zoom”). To simplify processing, we project hypergraphs into graph space using two different projections, each preserving specific characteristics. Generation is achieved by repeatedly applying the trained model to zoom in, gradually increasing detail until the desired hypergraph size is reached.
Preliminaries
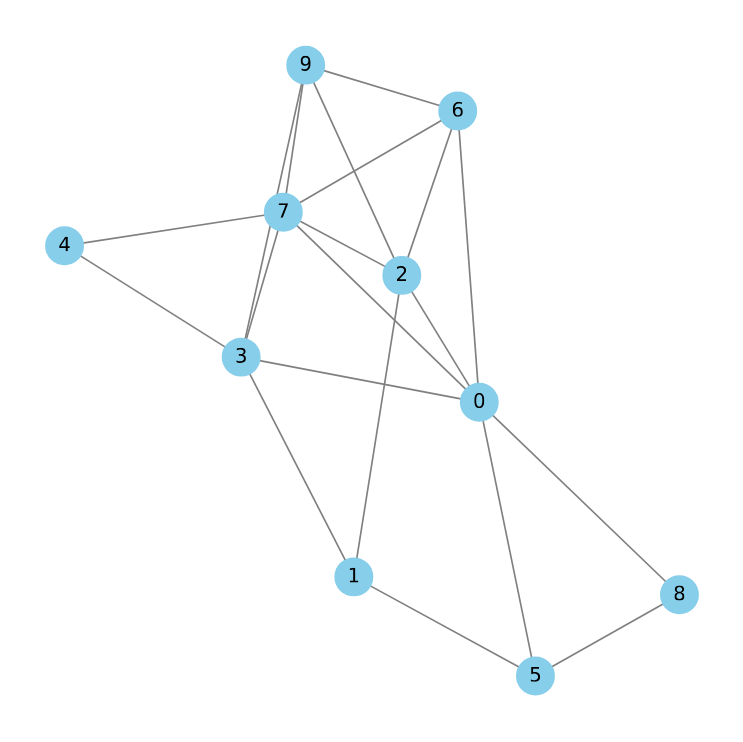
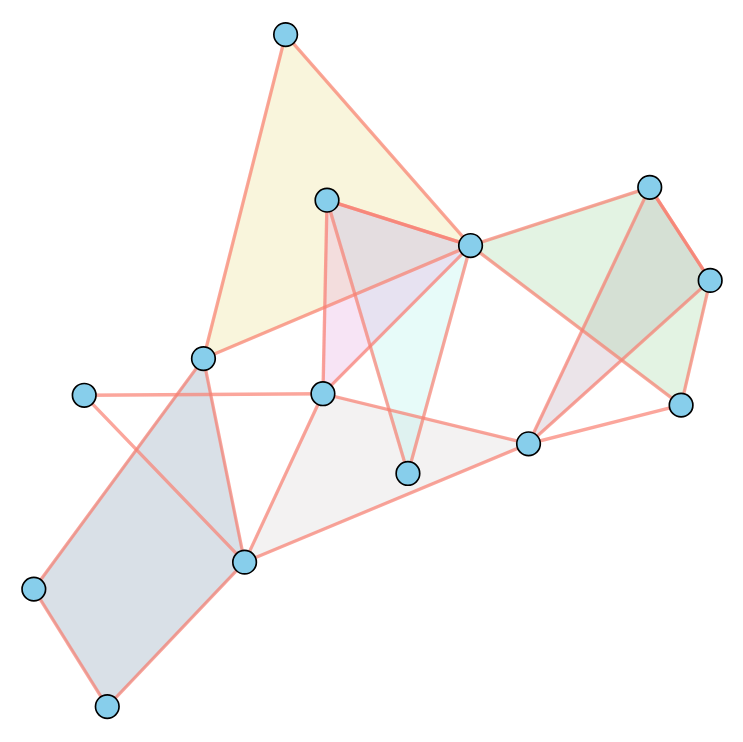
Traditional graphs consist of nodes connected by edges, with each edge linking precisely two nodes. Hypergraphs, however, offer a more versatile framework. In a hypergraph, edges — referred to as hyperedges — can connect an arbitrary number of nodes simultaneously. This generalization allows hypergraphs to capture higher-order and more nuanced relationships compared to standard graphs. This also makes them much more expressive, for instance:
- 3D meshes can be represented with triangles as 3-edges
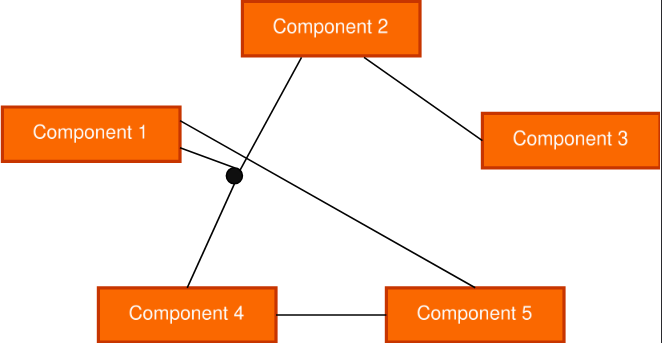
- Electronic circuits can be modeled with routers replaced by hyperedges connecting all associated components
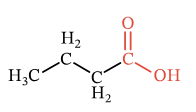
- Molecules can be depicted more accurately, with functional groups like carboxylic acids joined into hyperedges to highlight their higher-order relationship
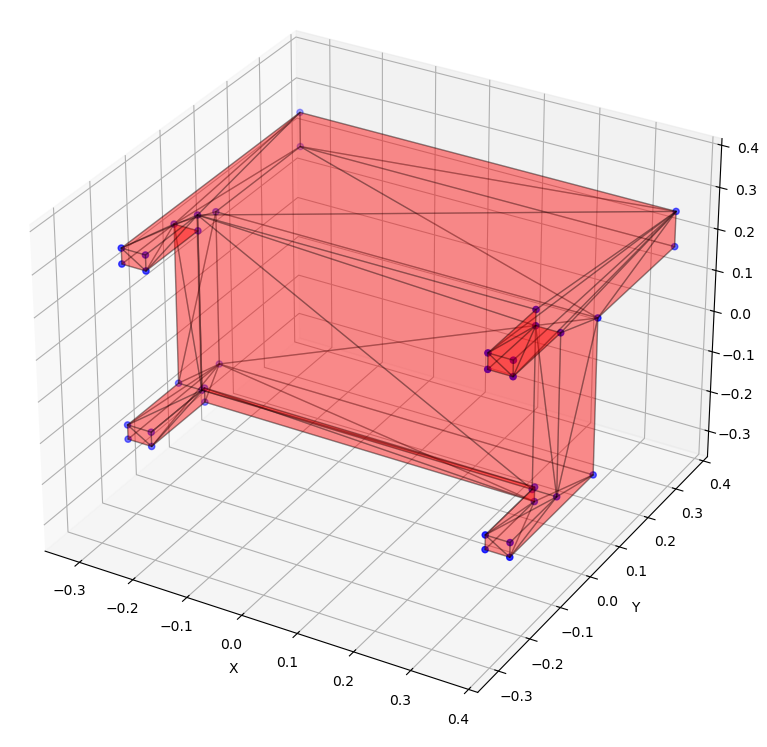
Our work aims to develop a deep-learning based method capable of generating new samples from a given distribution of hypergraphs. For instance, given examples of 3D meshes representing variants of an object, we want to train a model that can generate new instances. As an initial step, we focus solely on the underlying topology of hypergraphs, disregarding features of nodes and hyperedges.
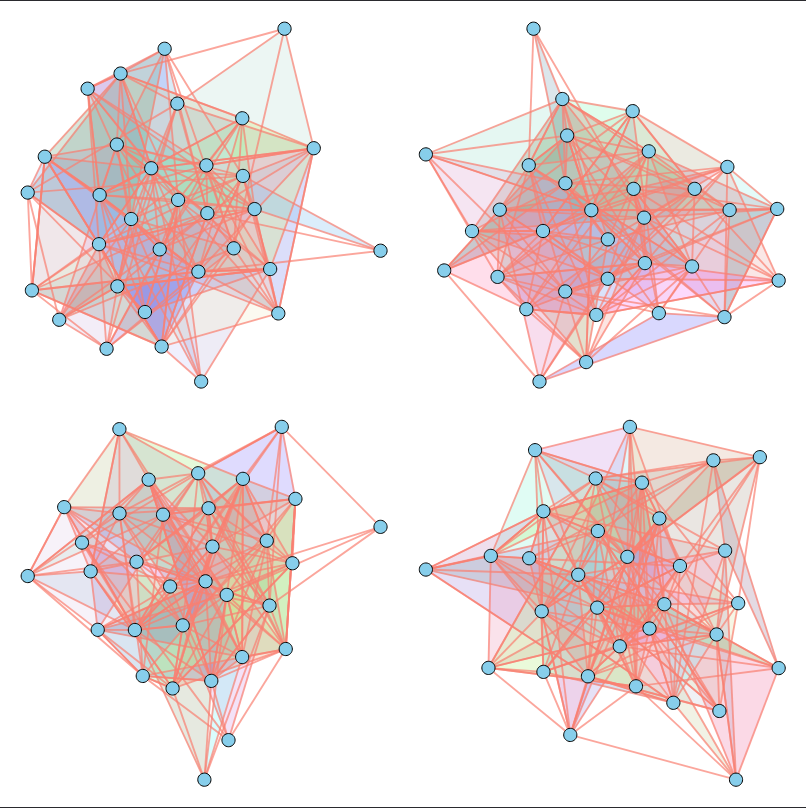
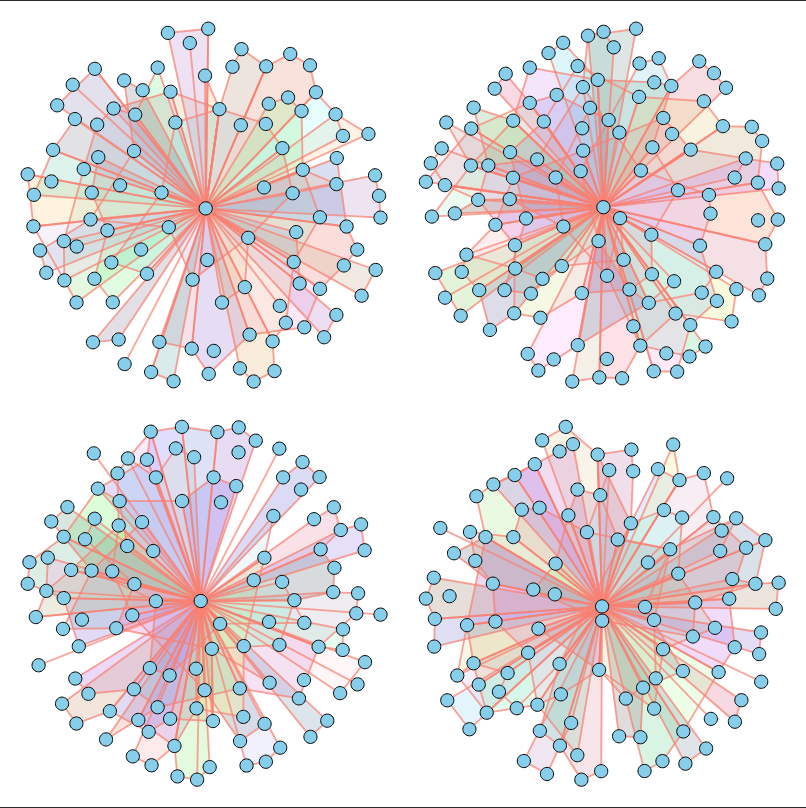
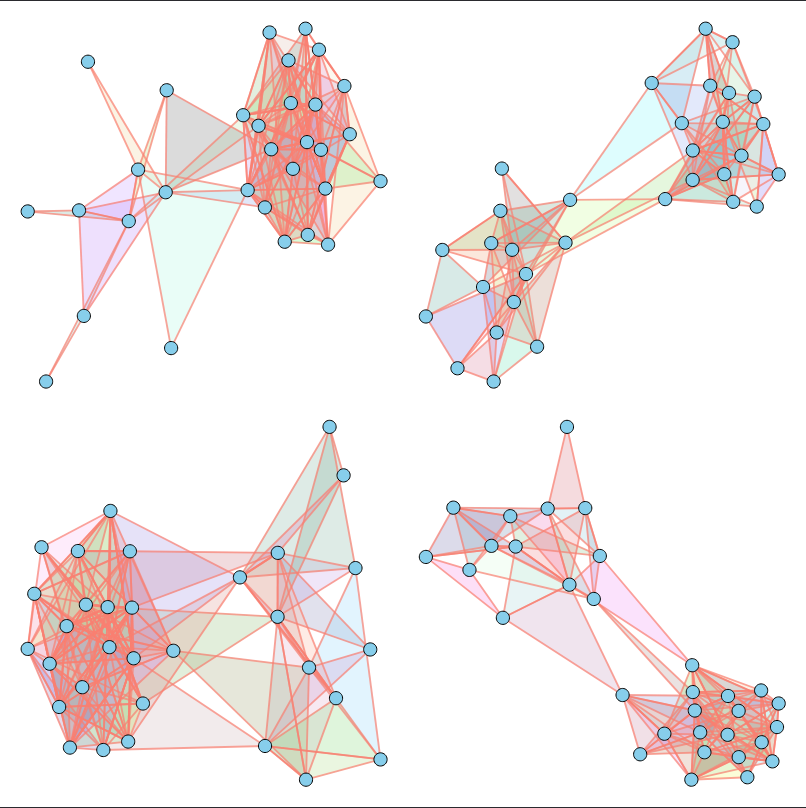
In other words, given a dataset of unfeatured hypergraphs (without 3D positions, component types, or other characteristics) that share similar topological properties, our goal is to train a model capable of generating new hypergraphs with matching properties.
Main Inspiration
Our work builds upon the foundation laid by the paper Efficient and Scalable Graph Generation through Iterative Local Expansion [2], which introduces an innovative method for graph generation. This approach draws inspiration from the tradition of graph coarsening algorithms.
When graphs become excessively large, certain algorithms may become computationally infeasible. To address this, it is possible to create smaller, representative versions of the original graph (called coarsened versions) that retain key characteristics and properties. The authors of the aforementioned paper employed a spectrum-preserving graph coarsening [3] technique. This method merges pairs of adjacent nodes in a way that preserves a subspace of the eigenvalues and eigenvectors of the original graph, maintaining crucial structural information.
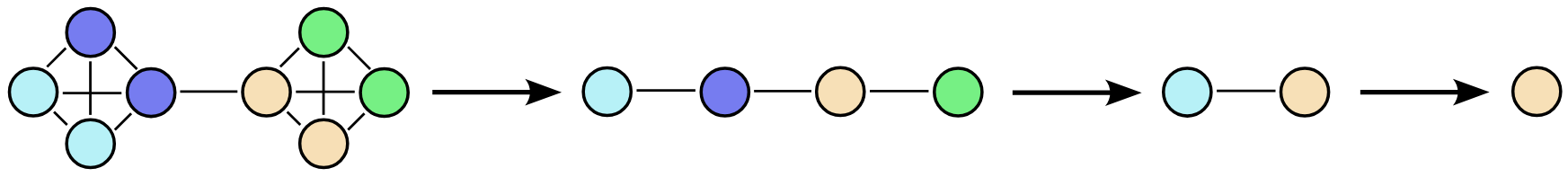
The coarsening algorithm is applied iteratively to create a coarsening sequence. The first element in this sequence is the original graph, while the last is a fully reduced version comprising a single node. The challenge then becomes training a model to invert this coarsening process.
Starting from a step in the sequence, the model must identify which nodes result from merging at the previous step. These nodes are then duplicated and connected, with each child retaining the connections of its parent node (expansion). The model then determines which edges should be retained in the final graph (refinement).
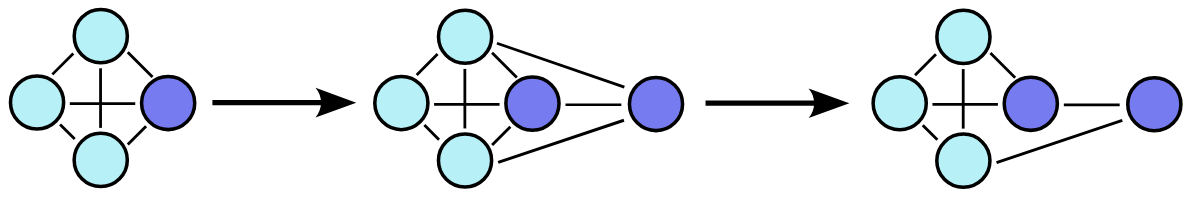
The generation process simply involves applying this expansion and refinement procedure iteratively until obtaining a graph of the desired size. This approach scales more efficiently compared to existing techniques, as it leverages the graph’s sparsity. When two parts of the graph become disconnected during an early generation step, there’s no need to check for connections later, significantly reducing computational overhead.
Our Work
We have successfully generalized this generation method to hypergraphs and tested it on several datasets. Given the challenges of working directly with hypergraphs due to variable hyperedge sizes and complex node relationships, we project the problem back into graph space using two equivalent representations.
Coarsening Sequence / Downsampling
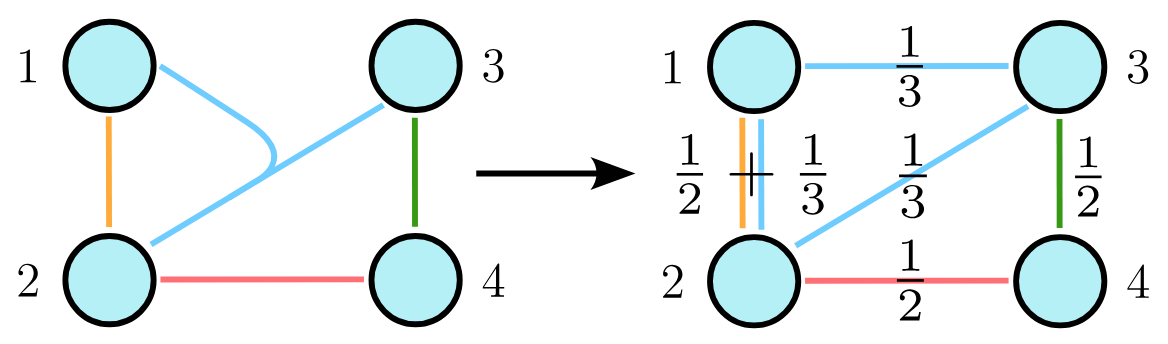
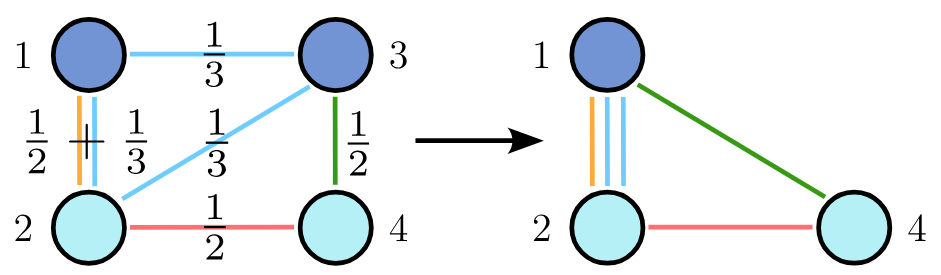
To apply coarsening algorithms to hypergraphs, we represent them as their clique expansion. In this representation, each hyperedge \(e\) is transformed into a clique weighted by \(1/ \vert e \vert\) , where \(\vert e \vert\) is the size of the hyperedge. When an edge appears in multiple cliques, its weight becomes the sum of the weights from each clique.
This clique expansion is particularly useful for coarsening, as it maintains the same number of nodes as the original hypergraph and shares its spectral properties. Consequently, node mergings that preserve spectral properties in the clique expansion also preserve them in the hypergraph. To construct a coarsening sequence for a given hypergraph, we create an equivalent sequence for its clique expansion and merge the same pairs of nodes.
However, it’s important to note that while deriving the clique expansion from a hypergraph is straightforward, the reverse process is NP-hard, which makes it infeasible to work with the clique expansion during generation.
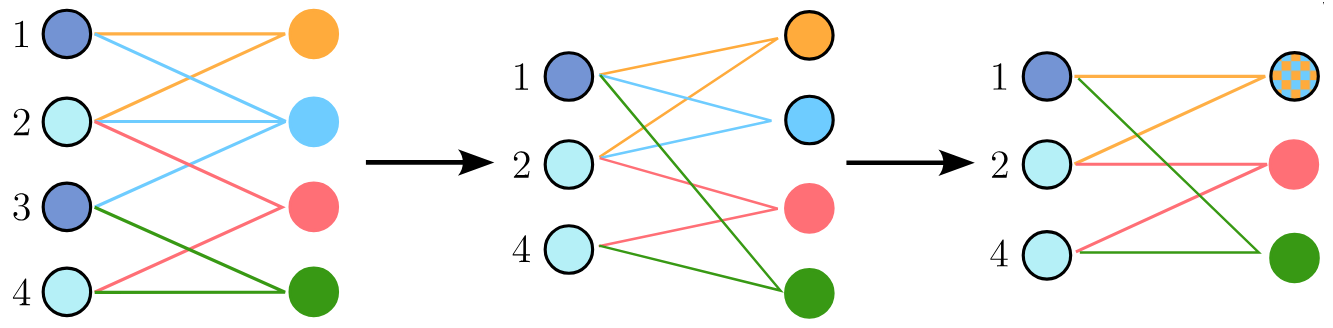
To address this limitation, we maintain another parallel representation of the hypergraph: the star expansion. This representation depicts the hypergraph as a bipartite graph, with one set of nodes corresponding to the hypergraph’s nodes and the other representing hyperedges. Each node is connected to the hyperedges containing it.
When nodes are merged in the clique expansion, the same nodes are merged in the star expansion. If two nodes representing hyperedges have identical connections (i.e., represent the same hyperedge), they are also merged after the node mergings in the clique expansion. Beware that in this scheme, a single pair of node mergings can result in the merging of up to three hyperedges.
Coarsening Inversion / Upsampling
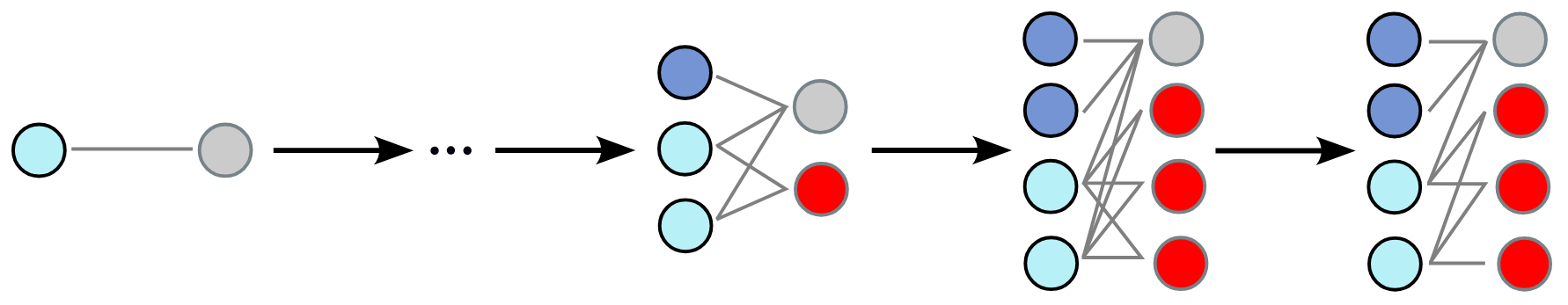
Beginning from a coarsening step, we train a model to identify which nodes and hyperedges result from merging at the previous coarsening step. These nodes are then expanded (nodes are duplicated twice, while hyperedges can be duplicated two or three times), with each child retaining its parent’s connections. Then the model is trained to selectively filter out regular edges, reconstructing an accurate topology. Generation starts with a single pair of connected node (the minimal bipartite graph) and iteratively applies the above scheme until obtaining a hypergraph of the desired size. Once it is attained, we can retrieve the generated hypergraph by replacing each node representing a hyperedge with a hyperedge connecting all its neighbors.
Pipeline
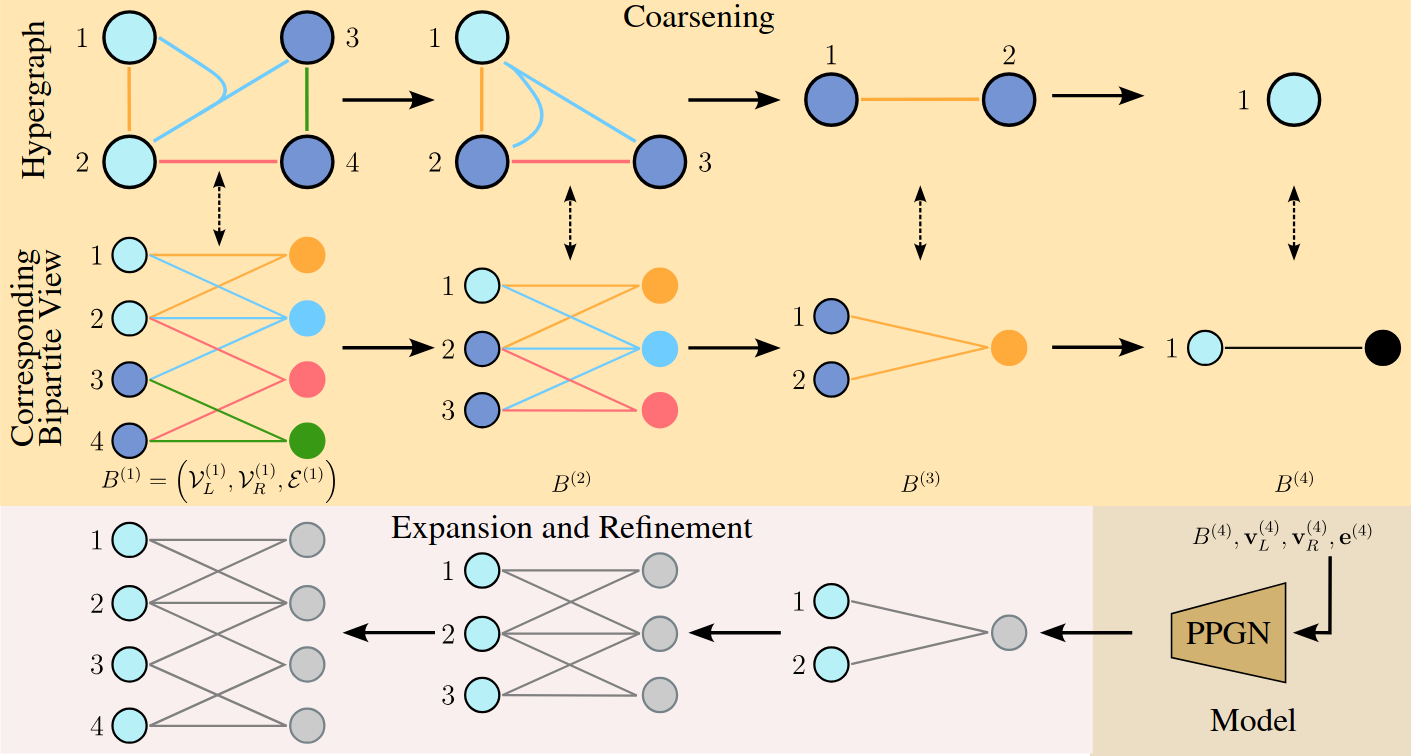
Results and Limitations
Results
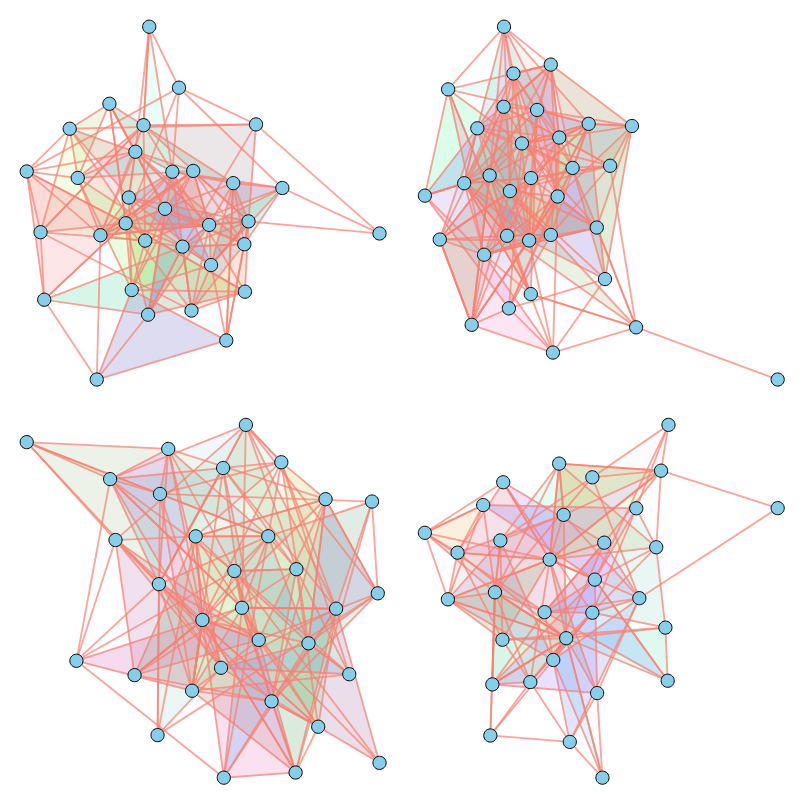
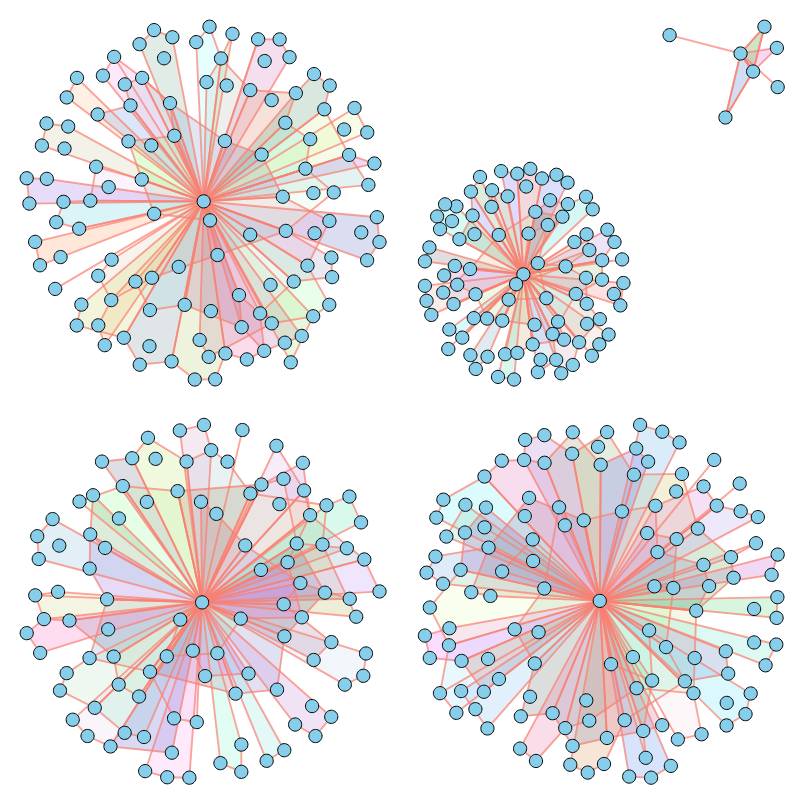
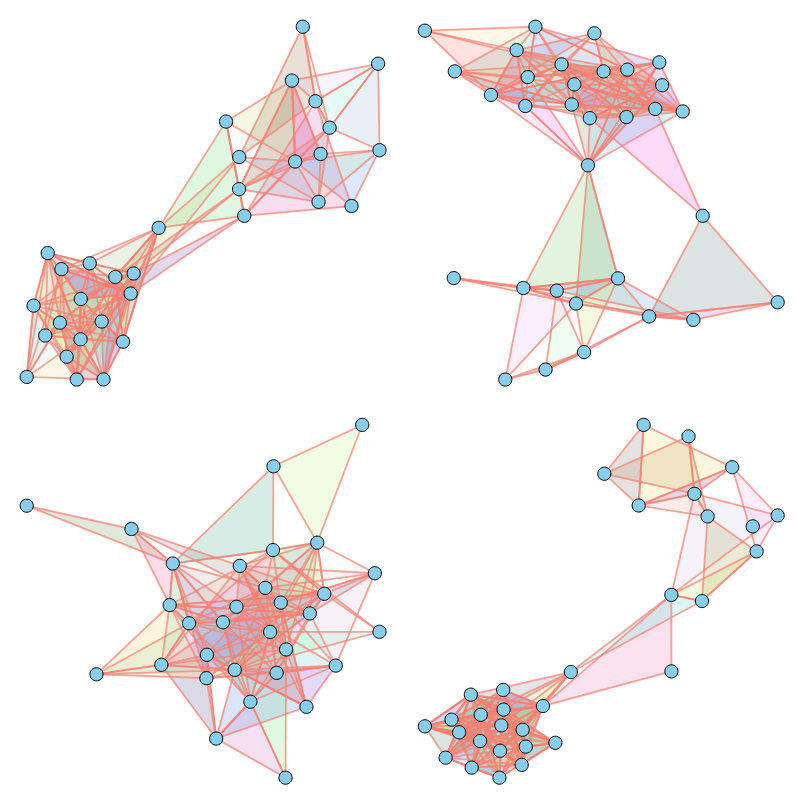
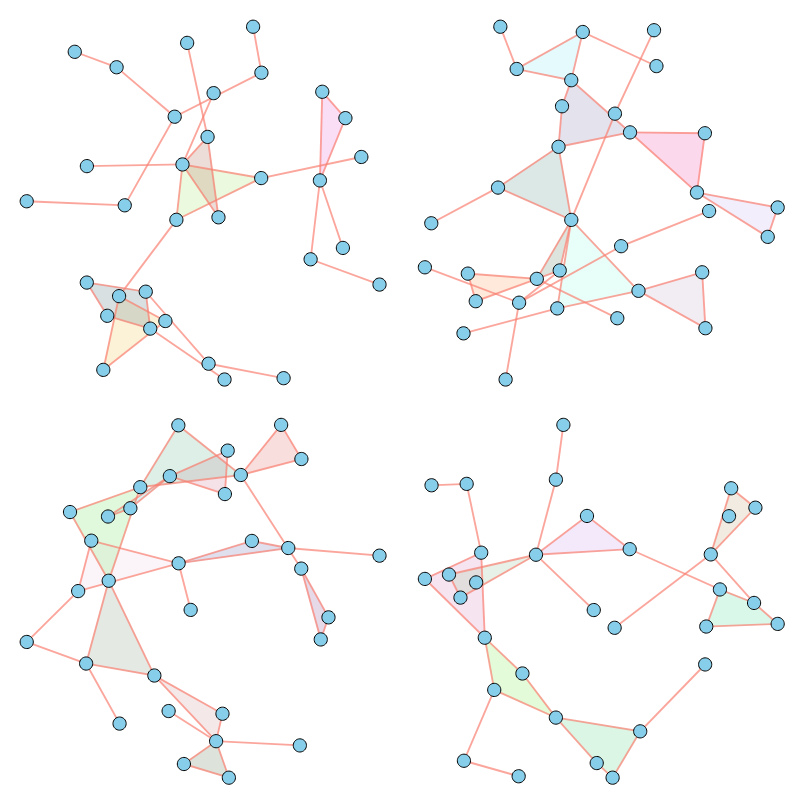
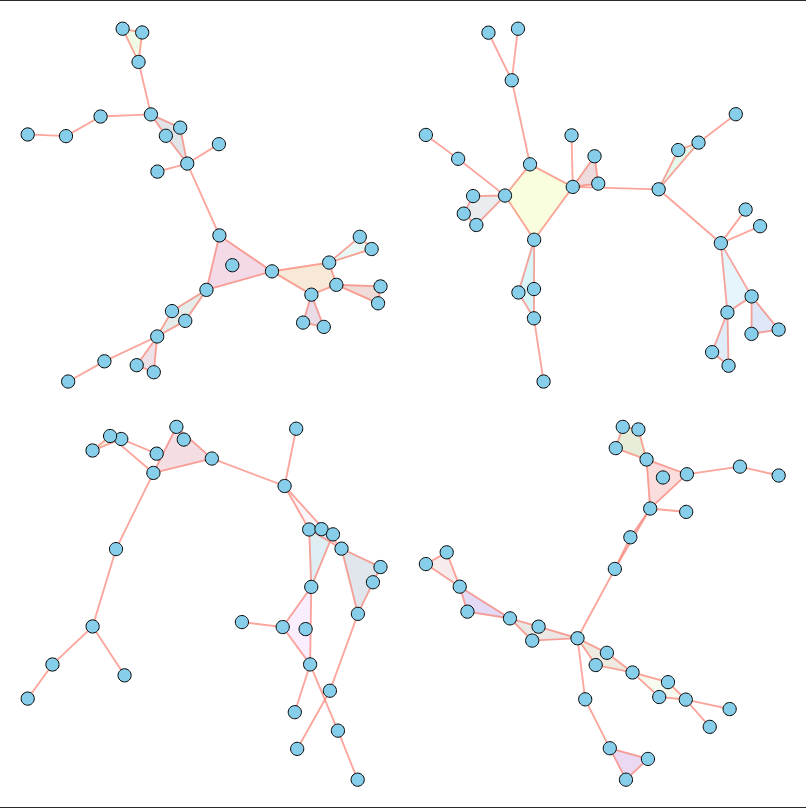
As illustrated above, our method successfully imitates a variety of properties. For a comprehensive analysis of numerical results across various metrics, please refer to our paper.
Limitations
Our method currently appears to disregard the specified number of nodes during generation, instead sampling from the node count distribution of the training data. Additionally, it encounters difficulties when dealing with datasets containing large hypergraphs.
Conclusion
This work represents a significant first step towards deep-learning based hypergraph generation. Our future research will focus on improving the model’s accuracy during generation, particularly concerning node count adherence, and expanding support for feature generation.
When using our work, please cite our paper:
@inproceedings{gailhard2025hygene,
title={HYGENE: A Diffusion-based Hypergraph Generation Method},
author={Gailhard, Dorian and Tartaglione, Enzo and Naviner, Lirida and Giraldo, Jhony H.},
booktitle={Proceedings of the AAAI Conference on Artificial Intelligence},
year={2025},
}
Acknowledgments
We acknowledge the ANR – FRANCE (French National Research Agency) for its financial support of the System On Chip Design leveraging Artificial Intelligence (SODA) project under grant ANR-23-IAS3-0004. This project has also been partially funded by the Hi!PARIS Center on Data Analytics and Artificial Intelligence.
References
- HYGENE: A Diffusion-based Hypergraph Generation Method [PDF]
Gailhard, D., Tartaglione, E., Naviner, L. and Giraldo, J., 2025, AAAI Conference on Artificial Intelligence. - Efficient and scalable graph generation through iterative local expansion [PDF]
Bergmeister, A., Martinkus, K., Perraudin, N. and Wattenhofer, R., 2024, International Conference on Learning Representations. - Graph reduction with spectral and cut guarantees [PDF]
Loukas, A., 2019, Journal of Machine Learning Research.